An Australian cancer researcher is raising the alarm about ‘paper mills’ that are churning out hundreds of fraudulent pre-clinical publications on cancer gene biomarker candidates.
Professor Jennifer Byrne of the Molecular Oncology Laboratory, The Children’s Hospital at Westmead, NSW, is one of a number of researchers who have identified a recurring pattern of manipulated or duplicate images and graphs being used in gene knockdown studies that target under-studied human genes.
The group believes the papers are being generated and sold by shady companies in China to doctors at institutions that require them to have scientific papers published in an international journal in order to get their MD or promotion
Working in conjunction with a group of ‘image forensic detectives’, Professor Byrne and colleagues have identified a series of 400 papers with suspiciously similar Western Blot figures, flow cytometry plots, bar graph patterns and even title structures.
The pattern of similarities from different authors in different institutions suggests that the papers are being prepared by the same source based on a ‘recipe’ or template with slight variations in each paper, says Professor Byrne, who is also Director Of Biobanking with NSW Health Pathology.
“We believe that features of the single-gene knockdown publications that we have studied could be consistent with a new and highly concerning form of systematic research fraud that employs a ‘theme and variations’ model for content generation,” they write in FEBS Letters.
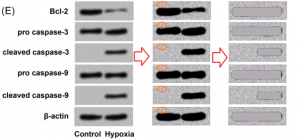 Professor Byrne says the paper mills appear to be taking advantage of the fact that there are 40,000 mostly unstudied genes to create a high output of papers describing nucleotide sequences that are of little value without a hypothesis, even if they are genuine. Many of the papers appear to be using manipulated images or stock images that bear no relation to the nucleotide sequences described in the text.
Professor Byrne says the paper mills appear to be taking advantage of the fact that there are 40,000 mostly unstudied genes to create a high output of papers describing nucleotide sequences that are of little value without a hypothesis, even if they are genuine. Many of the papers appear to be using manipulated images or stock images that bear no relation to the nucleotide sequences described in the text.
“The data presented in single-gene knockdown papers would be easy to fabricate at scale. Data shown in graphical format (eg changes in gene transcript levels, cell proliferation over time, and cell sorting results in response to gene knockdown) could be created by simply typing data into programmes such as Excel. The creation of figures showing digital images could also be achieved at scale using digital image banks, which could either be created by the paper mill or accessed externally,” they write.
And as well as being easy targets for fraudulent research, under-studied genes also make the peer review process difficult because few people have the expertise or experience to verify the results, they add.
Professor Byrnes says it’s likely there are thousands of such paper mill‐generated papers already concealed within the research literature and they will lead to cancer biomarker research to be misdirected.
“Such fraudulent publications both individually and collectively damage functional genomics research and therefore biomarker research, by generating incorrect information about gene function that appears to be relevant to human disease. This incorrect information will be most damaging where existing knowledge is limited, which will likely be the case for most of the genes that are deliberately targeted,” she wrote in a previous article in Biomarker Insights.
To counter the paper mill approach they propose that publishers introduce a system of rapidly published ‘Editorial Notes‘ to flag reported errors that should be investigated.
“Breaking the paper mill business model will also require journals, researchers, and institutions to both recognize and then reject or retract paper mill‐generated manuscripts and publications, such that they are stripped of all scientific and career value,” they conclude.