Understanding of IBD, its causes, prognosis and potential treatments will ultimately come from a ‘multi-omics’ approach, according to speakers at ECCO 2019.
The current ‘reductionist’ approaches that focus on single areas such as the microbiome or inflammatory responses fail to capture the interplay of systems that undoubtedly underlie the range of diseases that make up IBD, according to Professor Jonas Halfvarson of the Department of Gastroenterology, Faculty of Medicine and Health, Örebro University, Norway.
He said our current artificial classification of IBD into two main conditions Crohn’s Disease (CD) and ulcerative colitis (UC) may be a hindrance to understanding what is potentially a wide range of different diseases that have different causes, natural histories and different responses to the treatments.
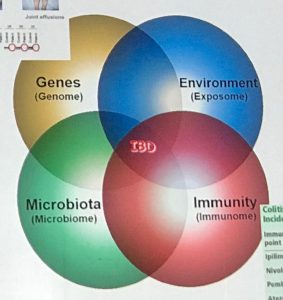 It is therefore essential to look at the wider picture of the contributions and interactions of the four ‘omes’ of the genome, environment (exposome), microbiome and immunity (the immunome’) he told the conference.
It is therefore essential to look at the wider picture of the contributions and interactions of the four ‘omes’ of the genome, environment (exposome), microbiome and immunity (the immunome’) he told the conference.
He cited the example of attempts to characterise the molecular biology of IBD, which had found more than 200 proteins from ‘IBD’ risk genes, some CD-specific, some specific for UC, but also wide overlap between these conditions and with other immune-related diseases.
This suggested that IBD was probably not a singular clinical entity but may consist of many sub-mechanisms that clinically give rise to a similar “IBD’ disease entity, he said.
Similarly with the microbiome, there was evidence of overlap between CD and UC microbiota, and also a recent finding that the microbiome of people with IBD may fluctuate in and out of the ‘healthy’ profile over time.
However, there was also recent work on DNA methylation profiles of children with IBD that suggested it may be possible to identify biomarkers of disease, and also genetic markers that predict response to treatment with drugs such as thiopurines.
And another recent study had shown that it was possible to use Multi-Omics Factor Analysis based on colonic and ileal biopsies and circulating CD14+ monocytes and CD4+ T cells from patients with active Crohn’s disease to predict response to ustekinumab therapy, he noted. A 10-feature transcriptomic and 15-feature genomic panel predicting endoscopic response to ustekinumab standard dosage, according to findings also presented at the ECCO 2019 meeting.
Therefore the vision for the future would be to have a multi-omics panel based on the microbiome, environmental factors, the genome and immune factors that would identify the disease, its prognosis and thus guide clinicians in clinical decisions and identify the right drug for the patient, he said.
“I think we have emerging data for multi-omics profiling of the molecular biology of IBD, even though we don’t have the answers today.”
“It might be that the current diagnostic criteria, classification system and how we define responses to therapy may hamper our attempts to identify profiles of IBD,” he said.